
- Cytoscape linux how to#
- Cytoscape linux install#
- Cytoscape linux software#
- Cytoscape linux code#
- Cytoscape linux download#
As a plug-in developer, you have to choose between something that works right now, but will go away eventually, or something that is clearly the future, but might take a long time to materialise. But Cytoscape 3 has been in the making for a long time. The upcoming third version promises to fix all the minor and major problems that exist under the hood. Cytoscape is a great tool as long as you don’t look too closely at what’s going on inside. Today I was fortunate enough to be attending the Cytoscape developer workshop at Vizbi 2012, where I learned a few new things.įirstly, one of my goals was to find out about the current state of Cytoscape development. It’s great because it’s so easy to create plug-ins. Substitute the dummy host (192.168.5.130) and port (8080) values for the appropriate values of your proxy.Ĭytoscape is a popular network visualisation and analysis tool. Instead, go to your Cytoscape installation directory, and look for a file named Cytoscape.vmoptions. It does not work for plug-ins (sorry, “apps”) that make use of off-the-shelf Java libraries. The problem is that it doesn’t work – it stores the proxy settings in a special way that only some bits of Cytoscape are aware of. If you are using Cytoscape 2.X or 3.0 behind a proxy, and you know your proxy settings, you may find the following useful.Ĭytoscape has a “proxy server settings” dialog, as described in the manual.
Cytoscape linux software#
This is usually done for you, but Bioinformaticians tend to use “non-standard” software that you’ll have to configure yourself. As a consequence, all the software on your computer needs to be configured to be proxy-aware. In large companies, you often find that direct web access is blocked: you have to ask a proxy server to request web pages on your behalf (The proxy also does stuff like scanning for viruses and malware). Posted in Uncategorized | Comments Closed Tags: cytoscape, semantic web, sparql, visualization
Cytoscape linux code#
What’s even better is that the source code is available on github.Īlso, if you have a chance, come see my poster at Vizbi 2015 in Boston.
Cytoscape linux download#
Or download it on from the app store website.
Cytoscape linux install#
Just install Cytoscape 3.0, start it, and go to menu->Apps->App Manager and search for “General SPARQL”. The easiest way to get the app is simply from the Cytoscape App manager. The app works with privately managed triple stores just as well. But as great as these resources are, public triple stores can sometimes be overloaded. The initial set is designed to work with public bioinformatics SPARQL endpoints provided by the EBI and Bio2RDF. When you first install the app, it comes pre-configured with a basic set of SPARQL queries, although it’s possible to provide your own set.
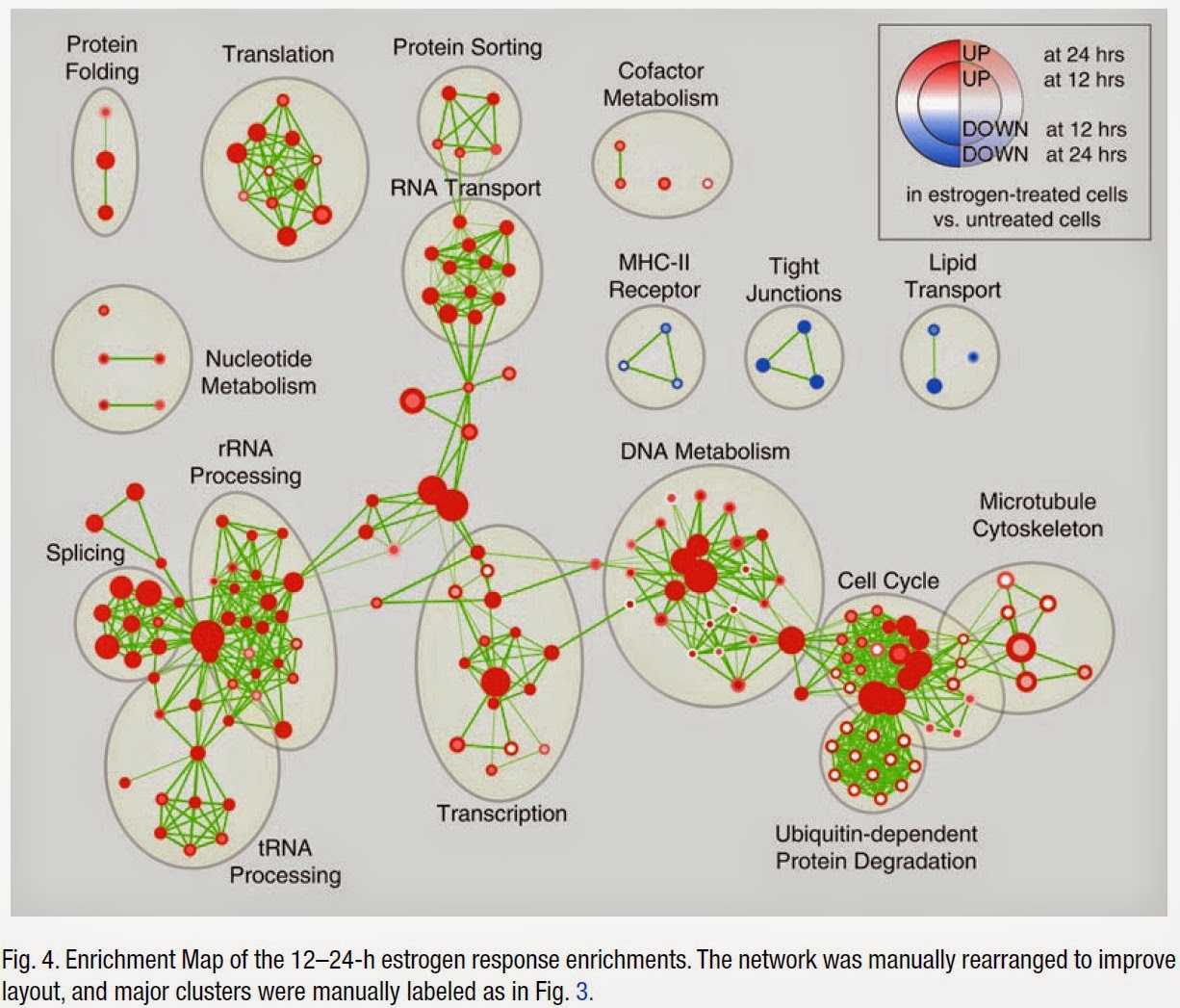
When you click on them, a query is sent off in the background, and the result is mapped to your network according to certain rules. Each item in the search menu, and each item in the context (right-click) menu, is backed by one SPARQL query. In the background, the General SPARQL app maintains a list of SPARQL queries. Watch this screencast and it will start to make sense: And you can continue this process, jumping from one entity to the next. Or the gene that encodes for your protein. Or all the reactions that your protein is part of. For example, all the pathways that are related to your protein. You can then right-click on this node to pull in related entities. The app places a single node in your network. Edges can represent any type of relation between those entities.įor example, you can start by searching for a protein of interest.
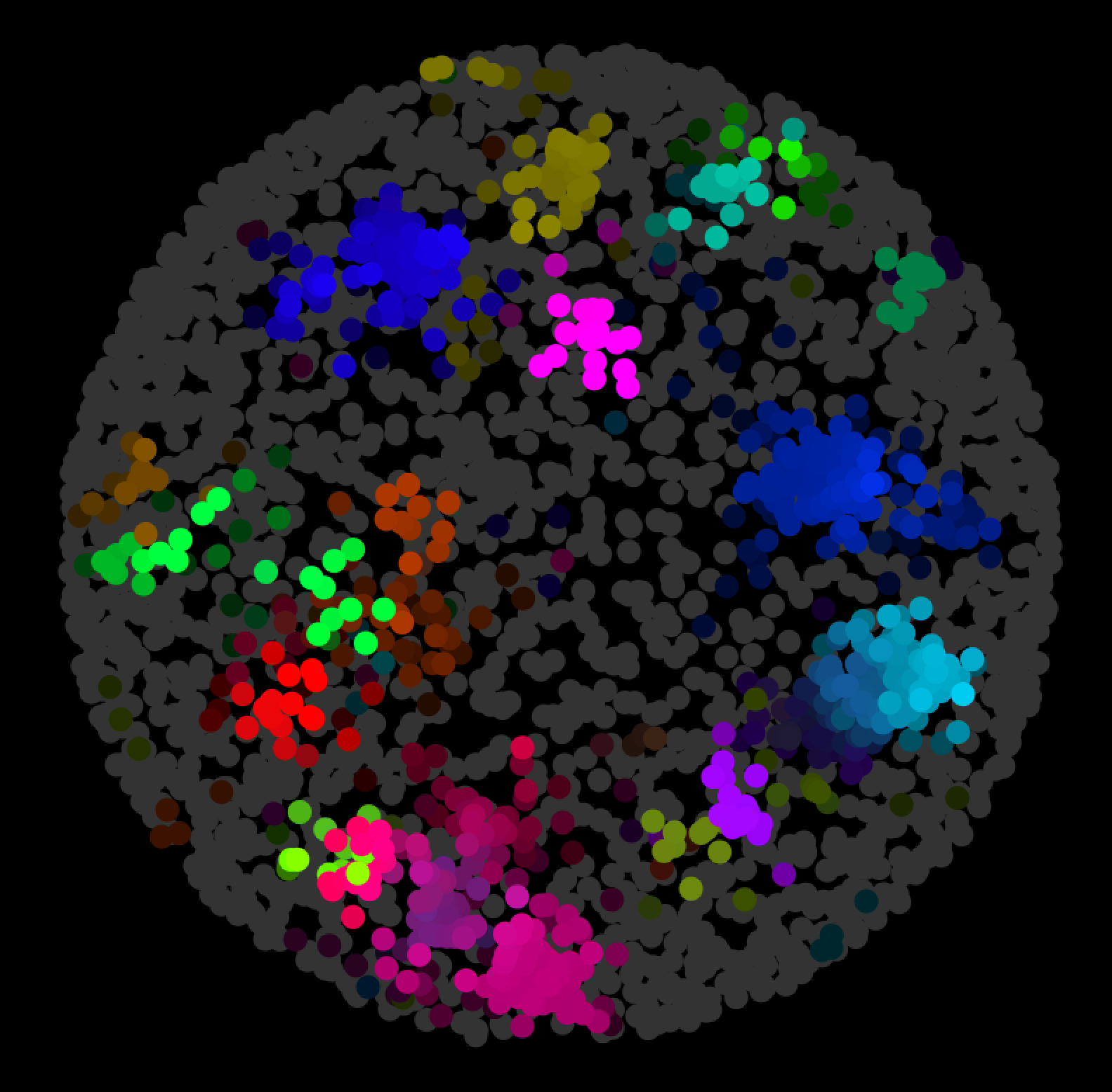
Nodes can represent various biological entities, such as: a pathway, a protein, a reaction, or a compound. Nodes and edges can be added to a network in a piecemeal fashion. The app lets you build a network step by step. The General SPARQL app is one of the new ways to present triple data.
We shouldn’t expect scientists to write SPARQL queries anymore than we expect them to carry adjustable pliers to a restroom visit. The problem is that SPARQL is supposed to be only a piece of plumbing at the bottom of a software stack. The problem isn’t that SPARQL is hard to use per se (it’s really rather plain and sensible). Integrating data is only half of the problem, we also have to present that data.
Cytoscape linux how to#
Triple stores are great for data integration, but you still have to figure out how to put that data in the hands of scientists. But how do we put that data in the hands of scientists?Īt General Bioinformatics we put data in triple stores, and use SPARQL to query that data. We can now easily solve the problem of bioinformatics data integration.


 0 kommentar(er)
0 kommentar(er)
